Part 4: Advanced Analyses
Overview
Teaching: 60 min
Exercises: 35 minQuestions
How can I carry out confirmatory factor analysis?
How can I compare two models?
Objectives
Prepare data for CFA.
Fit a one factor and two-factor model
Compare the models
In order to carry out CFA on our data, we need to prep it by calculating subscores. Again, we will be using a number of functions from the tidyverse. We will also be using the lavaan package. This is a package for latent variable modeling. It’s creators have a nice website with tutorials and resources.
#load packages
library(tidyverse)
library(lavaan)
library(semPlot)
# read data
sem_dat <- read_csv("data/placement_2.csv")
# compute subscores
sem_dat <- gather(sem_dat, item, correct, 5:69)
sem_dat <- separate(sem_dat, item, into = c("number", "skill", "type"),
sep = "_", remove = FALSE)
sum_scores <- group_by(sem_dat, ID, skill, type) %>%
summarise(total = sum(correct, na.rm = TRUE)) %>%
unite(skill_type, 2:3, sep = "_") %>%
spread(skill_type, total)
# some of the subscores are a bit limited in number of questions. combine
sum_scores <- mutate(sum_scores, list_global = list_mi + list_prag,
read_global = read_mi + read_inf + read_purp)
We will start by fitting a one-factor confirmatory model.
# Confirmatory Factor Analyses ----
# 1-factor model
## define the model
one <- '
# latent variable definitions
language =~ list_det + list_global + list_inf + read_det + read_voc +
read_torg + read_global'
## fit the model
fit_one <- cfa(one, data = sum_scores, missing = "fiml")
## plot the model
semPaths(fit_one, "std", layout = "tree", intercepts = FALSE, residuals = T, nDigits = 2,
label.cex = 1, edge.label.cex=.95, fade = FALSE)
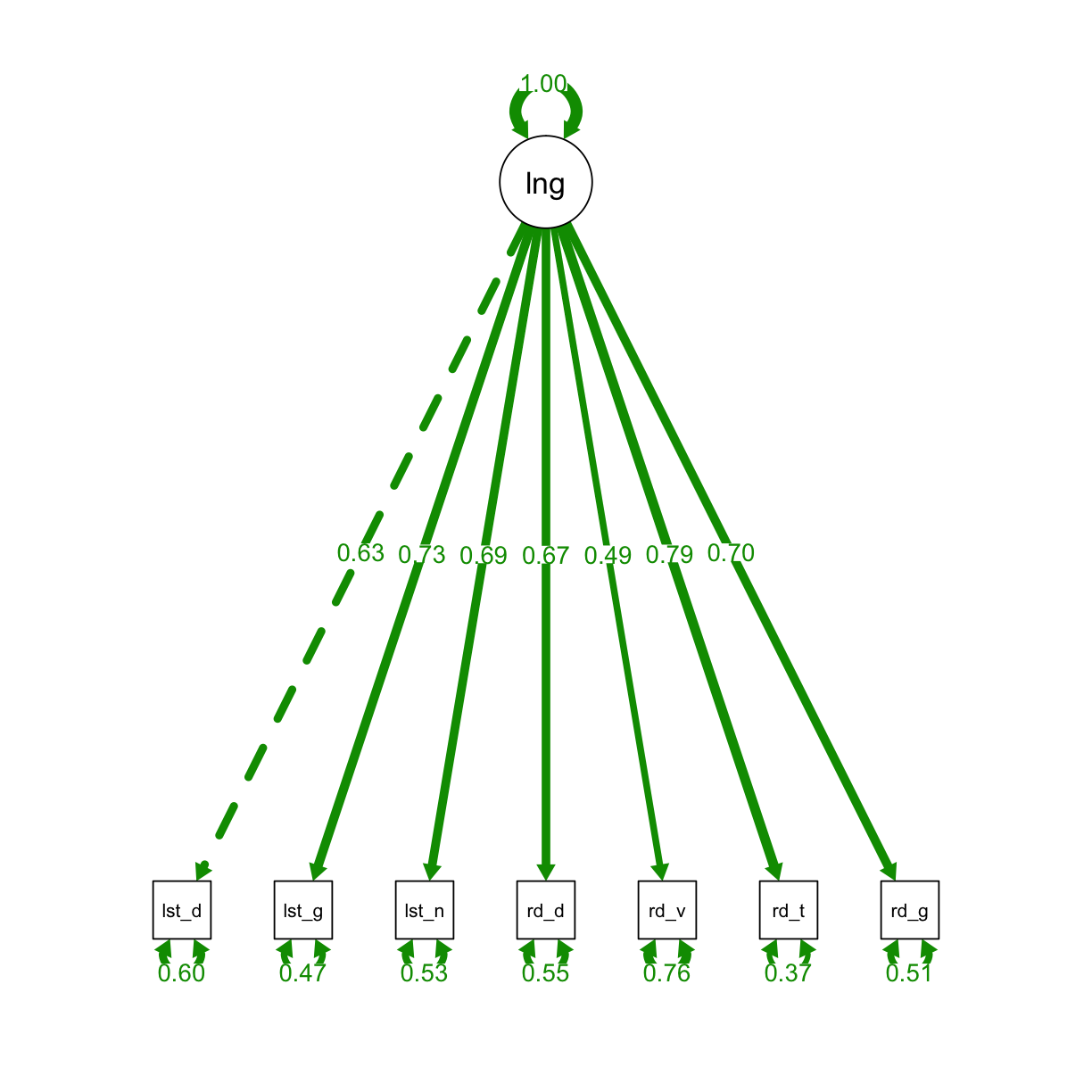
We can use summary to view the results, and we can check the diagnostics with resid and modindices.
#view results
lavaan::summary(fit_one, estimates = TRUE, standardized = TRUE, fit.measures = TRUE)
lavaan 0.6-3 ended normally after 25 iterations
Optimization method NLMINB
Number of free parameters 21
Number of observations 173
Number of missing patterns 1
Estimator ML
Model Fit Test Statistic 25.751
Degrees of freedom 14
P-value (Chi-square) 0.028
Model test baseline model:
Minimum Function Test Statistic 447.156
Degrees of freedom 21
P-value 0.000
User model versus baseline model:
Comparative Fit Index (CFI) 0.972
Tucker-Lewis Index (TLI) 0.959
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -2230.686
Loglikelihood unrestricted model (H1) -2217.811
Number of free parameters 21
Akaike (AIC) 4503.372
Bayesian (BIC) 4569.591
Sample-size adjusted Bayesian (BIC) 4503.094
Root Mean Square Error of Approximation:
RMSEA 0.070
90 Percent Confidence Interval 0.023 0.111
P-value RMSEA <= 0.05 0.201
Standardized Root Mean Square Residual:
SRMR 0.034
Parameter Estimates:
Information Observed
Observed information based on Hessian
Standard Errors Standard
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
language =~
list_det 1.000 1.206 0.631
list_global 1.216 0.154 7.893 0.000 1.467 0.728
list_inf 1.064 0.145 7.360 0.000 1.283 0.687
read_det 1.038 0.146 7.127 0.000 1.252 0.674
read_voc 0.458 0.081 5.658 0.000 0.553 0.492
read_torg 1.523 0.190 8.030 0.000 1.837 0.792
read_global 1.091 0.148 7.398 0.000 1.316 0.702
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.list_det 5.029 0.145 34.583 0.000 5.029 2.629
.list_global 5.237 0.153 34.164 0.000 5.237 2.597
.list_inf 3.740 0.142 26.341 0.000 3.740 2.003
.read_det 3.468 0.141 24.555 0.000 3.468 1.867
.read_voc 1.896 0.085 22.208 0.000 1.896 1.688
.read_torg 6.058 0.176 34.376 0.000 6.058 2.614
.read_global 3.214 0.143 22.529 0.000 3.214 1.713
language 0.000 0.000 0.000
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.list_det 2.203 0.266 8.273 0.000 2.203 0.602
.list_global 1.913 0.254 7.527 0.000 1.913 0.471
.list_inf 1.840 0.231 7.968 0.000 1.840 0.528
.read_det 1.884 0.235 8.027 0.000 1.884 0.546
.read_voc 0.955 0.108 8.836 0.000 0.955 0.757
.read_torg 1.999 0.294 6.796 0.000 1.999 0.372
.read_global 1.788 0.229 7.804 0.000 1.788 0.508
language 1.455 0.337 4.321 0.000 1.000 1.000
#view diagnostics
resid(fit_one, type="standardized")
$type
[1] "standardized"
$cov
lst_dt lst_gl lst_nf red_dt red_vc rd_trg rd_glb
list_det 0.000
list_global 1.919 0.000
list_inf -0.348 1.099 0.000
read_det -1.550 -1.913 -1.161 0.000
read_voc 0.952 0.173 -0.684 -0.083 0.000
read_torg -1.324 0.222 -0.729 1.139 -0.180 0.000
read_global -0.562 -3.020 1.036 1.585 -0.137 0.115 0.000
$mean
list_det list_global list_inf read_det read_voc read_torg
0 0 0 0 0 0
read_global
0
modindices(fit_one)
lhs op rhs mi epc sepc.lv sepc.all sepc.nox
24 list_det ~~ list_global 9.509 0.584 0.584 0.284 0.284
25 list_det ~~ list_inf 0.143 -0.068 -0.068 -0.034 -0.034
26 list_det ~~ read_det 2.132 -0.264 -0.264 -0.129 -0.129
27 list_det ~~ read_voc 1.079 0.125 0.125 0.086 0.086
28 list_det ~~ read_torg 1.856 -0.286 -0.286 -0.136 -0.136
29 list_det ~~ read_global 0.287 -0.096 -0.096 -0.048 -0.048
30 list_global ~~ list_inf 2.040 0.257 0.257 0.137 0.137
31 list_global ~~ read_det 3.046 -0.314 -0.314 -0.166 -0.166
32 list_global ~~ read_voc 0.035 0.022 0.022 0.016 0.016
33 list_global ~~ read_torg 0.065 0.055 0.055 0.028 0.028
34 list_global ~~ read_global 8.015 -0.509 -0.509 -0.275 -0.275
35 list_inf ~~ read_det 1.377 -0.200 -0.200 -0.108 -0.108
36 list_inf ~~ read_voc 0.452 -0.076 -0.076 -0.057 -0.057
37 list_inf ~~ read_torg 0.521 -0.146 -0.146 -0.076 -0.076
38 list_inf ~~ read_global 1.453 0.205 0.205 0.113 0.113
39 read_det ~~ read_voc 0.007 -0.010 -0.010 -0.007 -0.007
40 read_det ~~ read_torg 2.855 0.341 0.341 0.175 0.175
41 read_det ~~ read_global 5.295 0.391 0.391 0.213 0.213
42 read_voc ~~ read_torg 0.032 -0.023 -0.023 -0.017 -0.017
43 read_voc ~~ read_global 0.019 -0.015 -0.015 -0.012 -0.012
44 read_torg ~~ read_global 0.022 0.030 0.030 0.016 0.016
Now we will fit a two-factor model so that we can compare it to the one-factor model. We can use the same commands to examine the model.
# 2-factor model
## define the model
two <- '
# latent variable definitions
listening =~ list_det + list_global + list_inf
reading =~ read_det + read_voc + read_torg + read_global
#covariances
listening ~~ reading'
##fit the model
fit_two <- cfa(two, data = sum_scores, missing = "fiml")
##plot the model
semPaths(fit_two, "std", layout = "tree", intercepts = FALSE, residuals = TRUE, nDigits = 2,
label.cex = 1, edge.label.cex=.95, fade = FALSE)
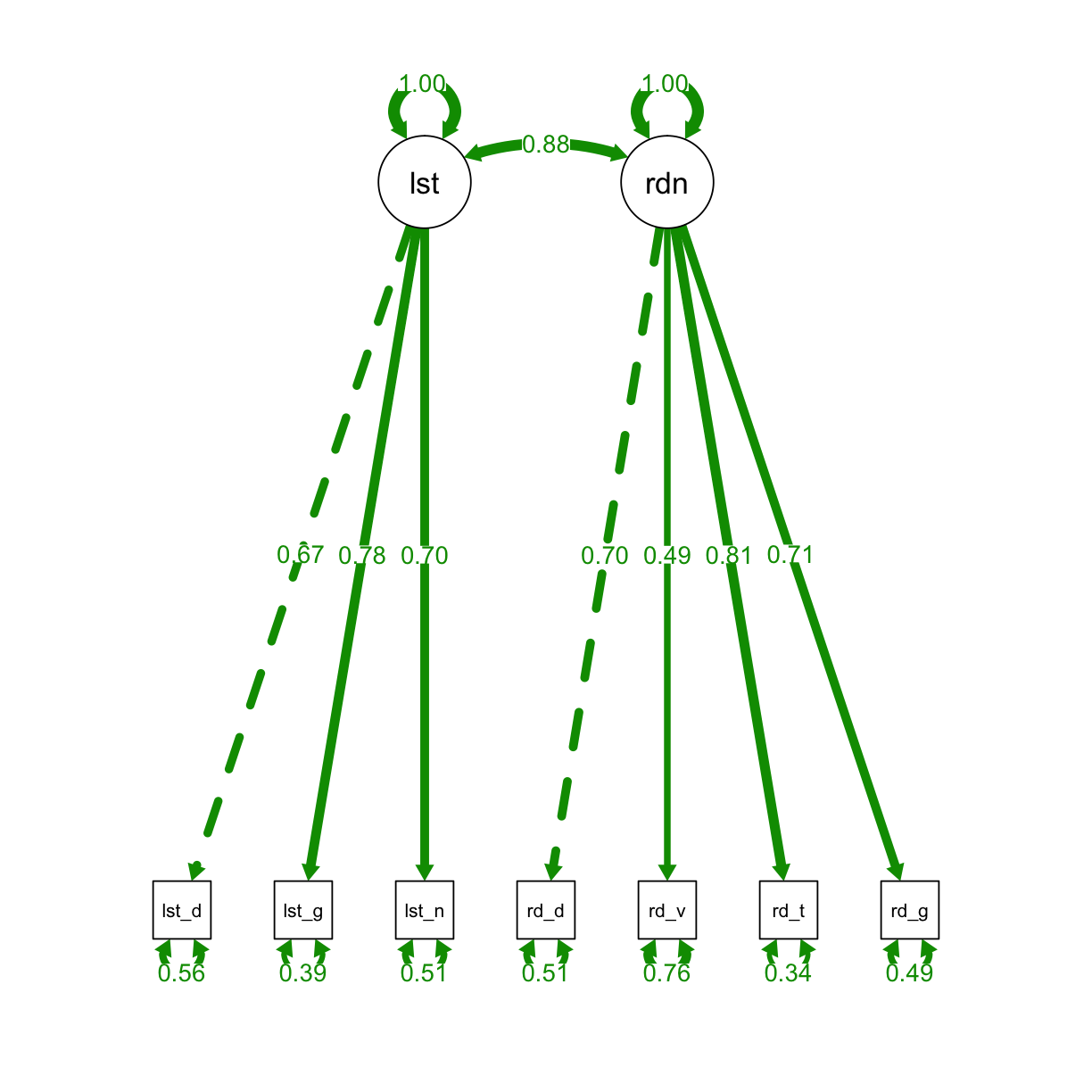
Exercise
Explore the model summary and check the diagnostics.
Solution
#view results lavaan::summary(fit_two, estimates = TRUE, standardized = TRUE, fit.measures = TRUE) #view diagnostics resid(fit_two, type="standardized") modindices(fit_two)
We can compare models with the anova function:
#compare models
anova(fit_one, fit_two)
Chi Square Difference Test
Df AIC BIC Chisq Chisq diff Df diff Pr(>Chisq)
fit_two 13 4496.3 4565.7 16.672
fit_one 14 4503.4 4569.6 25.751 9.0783 1 0.002587 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Now we will fit a model to that controls for method effects:
one_method <- '
# latent variable definitions
language =~ list_det + list_global + list_inf + read_det + read_voc +
read_torg + read_global
#listening method
list_det ~~ list_global
list_global ~~ list_inf
#reading method
read_det ~~ read_voc
read_det ~~ read_torg
read_det ~~ read_global
read_torg ~~ read_global'
## fit the model
fit_one_method <- cfa(one_method, data = sum_scores, missing = "fiml")
## plot the model
semPaths(fit_one_method, "std", layout = "tree", intercepts = FALSE, residuals = TRUE, nDigits = 2,
label.cex = 1, edge.label.cex=.95, fade = FALSE)
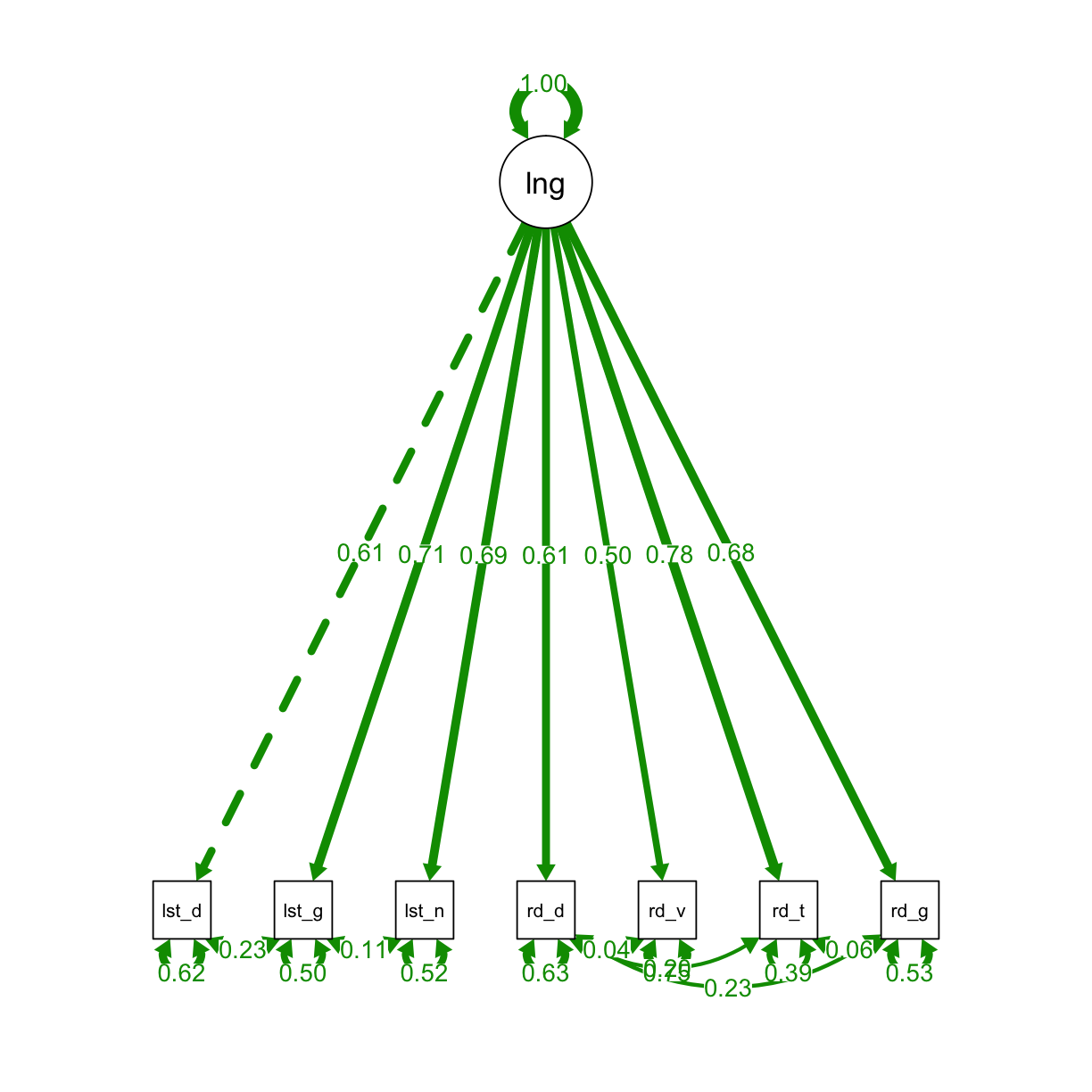
# view results
lavaan::summary(fit_one_method, estimates = TRUE, standardized = TRUE, fit.measures = TRUE)
lavaan 0.6-3 ended normally after 41 iterations
Optimization method NLMINB
Number of free parameters 27
Number of observations 173
Number of missing patterns 1
Estimator ML
Model Fit Test Statistic 7.629
Degrees of freedom 8
P-value (Chi-square) 0.471
Model test baseline model:
Minimum Function Test Statistic 447.156
Degrees of freedom 21
P-value 0.000
User model versus baseline model:
Comparative Fit Index (CFI) 1.000
Tucker-Lewis Index (TLI) 1.002
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -2221.625
Loglikelihood unrestricted model (H1) -2217.811
Number of free parameters 27
Akaike (AIC) 4497.251
Bayesian (BIC) 4582.390
Sample-size adjusted Bayesian (BIC) 4496.892
Root Mean Square Error of Approximation:
RMSEA 0.000
90 Percent Confidence Interval 0.000 0.086
P-value RMSEA <= 0.05 0.737
Standardized Root Mean Square Residual:
SRMR 0.018
Parameter Estimates:
Information Observed
Observed information based on Hessian
Standard Errors Standard
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
language =~
list_det 1.000 1.174 0.614
list_global 1.217 0.154 7.877 0.000 1.429 0.709
list_inf 1.101 0.162 6.779 0.000 1.293 0.692
read_det 0.966 0.172 5.627 0.000 1.135 0.611
read_voc 0.477 0.088 5.437 0.000 0.560 0.499
read_torg 1.540 0.230 6.693 0.000 1.809 0.780
read_global 1.091 0.180 6.045 0.000 1.281 0.683
Covariances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.list_det ~~
.list_global 0.491 0.232 2.114 0.035 0.491 0.228
.list_global ~~
.list_inf 0.202 0.209 0.965 0.335 0.202 0.105
.read_det ~~
.read_voc 0.052 0.115 0.454 0.650 0.052 0.036
.read_torg 0.430 0.280 1.538 0.124 0.430 0.202
.read_global 0.458 0.237 1.935 0.053 0.458 0.227
.read_torg ~~
.read_global 0.115 0.280 0.411 0.681 0.115 0.058
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.list_det 5.029 0.145 34.583 0.000 5.029 2.629
.list_global 5.237 0.153 34.160 0.000 5.237 2.597
.list_inf 3.740 0.142 26.341 0.000 3.740 2.003
.read_det 3.468 0.141 24.554 0.000 3.468 1.867
.read_voc 1.896 0.085 22.208 0.000 1.896 1.688
.read_torg 6.058 0.176 34.376 0.000 6.058 2.614
.read_global 3.214 0.143 22.529 0.000 3.214 1.713
language 0.000 0.000 0.000
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.list_det 2.279 0.295 7.720 0.000 2.279 0.623
.list_global 2.025 0.329 6.147 0.000 2.025 0.498
.list_inf 1.816 0.261 6.964 0.000 1.816 0.521
.read_det 2.164 0.306 7.075 0.000 2.164 0.627
.read_voc 0.947 0.110 8.609 0.000 0.947 0.751
.read_torg 2.101 0.411 5.113 0.000 2.101 0.391
.read_global 1.879 0.301 6.237 0.000 1.879 0.534
language 1.379 0.349 3.952 0.000 1.000 1.000
# view diagnostics
resid(fit_one_method, type="standardized")
$type
[1] "standardized"
$cov
lst_dt lst_gl lst_nf red_dt red_vc rd_trg rd_glb
list_det 0.000
list_global -0.058 -0.016
list_inf -0.208 -0.133 0.000
read_det -0.126 0.130 0.021 -0.065
read_voc 1.079 0.336 -0.992 -0.134 0.000
read_torg -0.710 1.322 -0.652 -0.034 -0.173 0.000
read_global 0.133 -2.101 1.459 -0.003 -0.022 0.000 0.000
$mean
list_det list_global list_inf read_det read_voc read_torg
0 0 0 0 0 0
read_global
0
modindices(fit_one_method)
lhs op rhs mi epc sepc.lv sepc.all sepc.nox
30 list_det ~~ list_inf 0.043 -0.054 -0.054 -0.026 -0.026
31 list_det ~~ read_det 0.023 -0.026 -0.026 -0.012 -0.012
32 list_det ~~ read_voc 1.155 0.129 0.129 0.088 0.088
33 list_det ~~ read_torg 0.862 -0.207 -0.207 -0.095 -0.095
34 list_det ~~ read_global 0.401 0.114 0.114 0.055 0.055
35 list_global ~~ read_det 0.105 0.057 0.057 0.027 0.027
36 list_global ~~ read_voc 0.015 0.015 0.015 0.011 0.011
37 list_global ~~ read_torg 2.708 0.378 0.378 0.183 0.183
38 list_global ~~ read_global 4.857 -0.399 -0.399 -0.204 -0.204
39 list_inf ~~ read_det 0.053 -0.042 -0.042 -0.021 -0.021
40 list_inf ~~ read_voc 0.831 -0.110 -0.110 -0.084 -0.084
41 list_inf ~~ read_torg 0.725 -0.198 -0.198 -0.102 -0.102
42 list_inf ~~ read_global 3.656 0.354 0.354 0.192 0.192
43 read_voc ~~ read_torg 0.029 -0.026 -0.026 -0.019 -0.019
44 read_voc ~~ read_global 0.000 -0.002 -0.002 -0.001 -0.001
Exercise
Compare the
one_fit_methodmodel with thefit_oneandfit_twomodels.Solution
## compare models anova(fit_one, fit_one_method) #yes, accounting for method effects is important anova(fit_two, fit_one_method) #inconclusive!
Key Points
lavaanandsemPlotare two useful packages for CFA.